Tutorial 5: Co-occurrence analysis
Andreas Niekler, Gregor Wiedemann
2017-09-11
This exercise will demonstrate how to perform co-occurrence analysis with R and the tm-package. It is shown how different significance measures can be used to extract semantic links between words.
Change to your working directory, create a new R script, load the tm-package and define a few already known default variables.
options(stringsAsFactors = FALSE)
library(tm)
textdata <- read.csv("data/sotu.csv", sep = ";", encoding = "UTF-8")
english_stopwords <- readLines("resources/stopwords_en.txt", encoding = "UTF-8")
# Create corpus object
m <- list(ID = "id", content = "text", DateTimeStamp = "date")
myReader <- readTabular(mapping = m)
corpus <- Corpus(DataframeSource(textdata), readerControl = list(reader = myReader))1 Sentence detection
The separation of the text into semantic analysis units is important for co-occurrence analysis. Context windows can be for instance documents, paragraphs or sentences or neighbouring words. One of the most frequently used context window is the sentence.
Documents are decomposed into sentences. Senteces are defined as a separate (quasi-)documents in a new corpus object of the tm-package. The further application of the tm-package functions remains the same. In contrast to previous exercises, however, we now use sentences which are stored as individual documents in the body.
Important: The sentence segmentation must take place before the other preprocessing steps because the sentence-segmentation-model relies on intact word forms and punctuation marks.
The following code defines two functions. One selects documents, the other decomposes a given document text into sentences with the help of a probabilistic model.
require(openNLP)
# Function to convert a document in a vector of sentences
convert_text_to_sentences <- function(text, lang = "en", SentModel = "resources/en-sent.bin") {
# Calculate sentenve boundaries as annotation with Apache OpenNLP Maxent-sentence-detector.
sentence_token_annotator <- Maxent_Sent_Token_Annotator(language = lang, model = SentModel)
# Convert text to NLP string
text <- NLP::as.String(text)
# Annotate the sentence boundaries
sentenceBoundaries <- NLP::annotate(text, sentence_token_annotator)
# Select sentences as rows of a new matrix
sentences <- text[sentenceBoundaries]
# return the sentences
return(sentences)
}# Function to convert a corpus of documents into a corpus of single sentences from documents
reshape_corpus <- function(currentCorpus, ...) {
# Extraction of all sentences from the corpus as a list
text <- lapply(currentCorpus, as.character)
# convert the text
pb <- txtProgressBar(min=0, max=length(text))
i <- 0
docs <- lapply(text, FUN=function(x){
i <<- i + 1
setTxtProgressBar(pb, i)
convert_text_to_sentences(x)
}, ...)
close(pb)
docs <- as.vector(unlist(docs))
# Create a new corpus of the segmented sentences
newCorpus <- Corpus(VectorSource(docs))
return(newCorpus)
}We apply the function reshape_corpus on our corpus of full speeches to receive a corpus of sentences.
# original corpus length and its first document
length(corpus)## [1] 231substr(as.character(corpus[[1]]), 0, 200)## [1] "Fellow-Citizens of the Senate and House of Representatives:\n\nI embrace with great satisfaction the opportunity which now presents itself\nof congratulating you on the present favorable prospects of our"# reshape into sentences
sentenceCorpus <- reshape_corpus(corpus)## ===========================================================================# reshaped corpus length and its first 'document'
length(sentenceCorpus)## [1] 62517as.character(sentenceCorpus[[1]])## [1] "Fellow-Citizens of the Senate and House of Representatives:\n\nI embrace with great satisfaction the opportunity which now presents itself\nof congratulating you on the present favorable prospects of our public\naffairs."as.character(sentenceCorpus[[2]])## [1] "The recent accession of the important state of North Carolina to\nthe Constitution of the United States (of which official information has\nbeen received), the rising credit and respectability of our country, the\ngeneral and increasing good will toward the government of the Union, and\nthe concord, peace, and plenty with which we are blessed are circumstances\nauspicious in an eminent degree to our national prosperity."CAUTION: The newly decomposed corpus has now reached a considerable size of 62517 sentences. Older computers may get in trouble because of insufficient memory during this preprocessing step.
Now we are returning to our usual pre-processing chain and apply it on the separated sentences.
# Preprocessing chain
sentenceCorpus <- tm_map(sentenceCorpus, removePunctuation, preserve_intra_word_dashes = TRUE)
sentenceCorpus <- tm_map(sentenceCorpus, removeNumbers)
sentenceCorpus <- tm_map(sentenceCorpus, content_transformer(tolower))
sentenceCorpus <- tm_map(sentenceCorpus, removeWords, english_stopwords)
sentenceCorpus <- tm_map(sentenceCorpus, stripWhitespace)Again, we create a document-term-matrix. Only word forms which occur at least 10 times should be taken into account. An upper limit is not set (Inf = infinite).
Additionally, we are interested in the joint occurrence of words in a sentence. For this, we do not need the exact count of how often the terms occur, but only the information whether they occur together or not. This can be encoded in a binary document-term-matrix. The parameter weighting in the control options calls the weightBin function. This writes a 1 into the DTM if the term is contained in a sentence and 0 if not.
minimumFrequency <- 10
binDTM <- DocumentTermMatrix(sentenceCorpus, control=list(bounds = list(global=c(minimumFrequency, Inf)), weighting = weightBin))2 Counting co-occurrences
The counting of the joint word occurrence is easily possible via a matrix multiplication (https://en.wikipedia.org/wiki/Matrix_multiplication) on the binary DTM. For this purpose, the transposed matrix (dimensions: nTypes x nDocs) is multiplied by the original matrix (nDocs x nTypes), which as a result encodes a term-term matrix (dimensions: nTypes x nTypes).
However, since we are working on very large matrices and the sparse matrix format (slam) which is used by the tm-package does not fully support the matrix multiplication, we first have to convert the binDTM into the format of the Matrix package which is more convenient to use.
# Convert to sparseMatrix matrix
require(Matrix)
binDTM <- sparseMatrix(i = binDTM$i, j = binDTM$j, x = binDTM$v, dims = c(binDTM$nrow, binDTM$ncol), dimnames = dimnames(binDTM))
# Matrix multiplication for cooccurrence counts
coocCounts <- t(binDTM) %*% binDTMLet’s look at a snippet of the result. The matrix has nTerms rows and columns and is symmetric. Each cell contains the number of joint occurrences. In the diagonal, the frequencies of single occurrences of each term are encoded.
as.matrix(coocCounts[202:205, 202:205])## advocating affair affairs affect
## advocating 11 0 0 0
## affair 0 18 1 0
## affairs 0 1 515 1
## affect 0 0 1 99Interprete as follows: affairs appears together 1 times with affect in the 62517 sentences of the SUTO addresses. affairs alone occurs 515 times.
3 Statistical significance
In order to not only count joint occurrence we have to determine their significance. Different significance-measures can be used. We need also various counts to calculate the significance of the joint occurrence of a term i (coocTerm) with any other term j: * k - Number of all context units in the corpus * ki - Number of occurrences of coocTerm * kj - Number of occurrences of comparison term j * kij - Number of joint occurrences of coocTerm and j
These quantities can be calculated for any term coocTerm as follows:
coocTerm <- "spain"
k <- nrow(binDTM)
ki <- sum(binDTM[, coocTerm])
kj <- colSums(binDTM)
names(kj) <- colnames(binDTM)
kij <- coocCounts[coocTerm, ]An implementation in R for Mutual Information, Dice, and Log-Likelihood may look like this. At the end of each formula, the result is sorted so that the most significant co-occurrences are at the first ranks of the list.
########## MI: log(k*kij / (ki * kj) ########
mutualInformationSig <- log(k * kij / (ki * kj))
mutualInformationSig <- mutualInformationSig[order(mutualInformationSig, decreasing = TRUE)]
########## DICE: 2 X&Y / X + Y ##############
dicesig <- 2 * kij / (ki + kj)
dicesig <- dicesig[order(dicesig, decreasing=TRUE)]
########## Log Likelihood ###################
logsig <- 2 * ((k * log(k)) - (ki * log(ki)) - (kj * log(kj)) + (kij * log(kij))
+ (k - ki - kj + kij) * log(k - ki - kj + kij)
+ (ki - kij) * log(ki - kij) + (kj - kij) * log(kj - kij)
- (k - ki) * log(k - ki) - (k - kj) * log(k - kj))
logsig <- logsig[order(logsig, decreasing=T)]The result of the four variants for the statistical extraction of co-occurrence terms is shown in a data frame below. It can be seen that frequency is a bad indicator of meaning constitution. Mutual information emphasizes rather rare events in the data. Dice and Log-likelihood yield very well interpretable contexts.
# Put all significance statistics in one Data-Frame
resultOverView <- data.frame(
names(sort(kij, decreasing=T)[1:10]), sort(kij, decreasing=T)[1:10],
names(mutualInformationSig[1:10]), mutualInformationSig[1:10],
names(dicesig[1:10]), dicesig[1:10],
names(logsig[1:10]), logsig[1:10],
row.names = NULL)
colnames(resultOverView) <- c("Freq-terms", "Freq", "MI-terms", "MI", "Dice-Terms", "Dice", "LL-Terms", "LL")
print(resultOverView)## Freq-terms Freq MI-terms MI Dice-Terms Dice LL-Terms
## 1 spain 439 spain 4.958694 spain 1.00000000 cuba
## 2 states 145 amistad 4.042404 cuba 0.13915416 united
## 3 united 136 antilles 4.042404 spanish 0.11550152 states
## 4 government 112 madrid 3.900087 france 0.08205128 spanish
## 5 treaty 55 catholic 3.893984 island 0.07235890 treaty
## 6 cuba 51 ton 3.754722 madrid 0.06967213 france
## 7 made 46 subdue 3.734919 treaty 0.06944444 madrid
## 8 war 45 queen 3.572400 florida 0.06106870 island
## 9 part 44 cubans 3.511775 friendly 0.05963303 government
## 10 relations 39 disavowed 3.492357 colonies 0.05936920 florida
## LL
## 1 243.67705
## 2 226.06445
## 3 195.38370
## 4 180.45952
## 5 124.91273
## 6 111.44823
## 7 106.42963
## 8 89.43827
## 9 86.72616
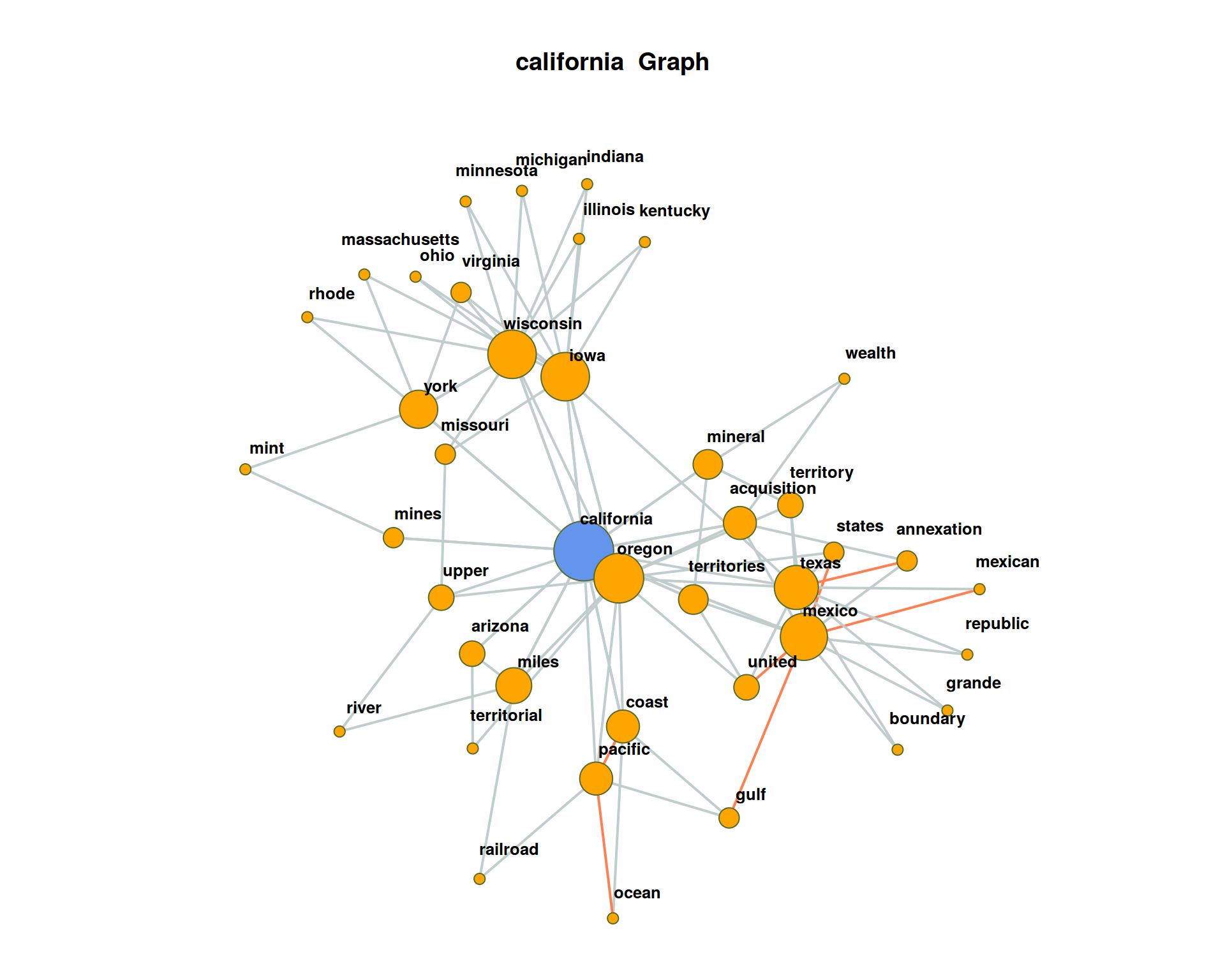
## 10 77.980974 Visualization of co-occurrence
In the following, we create a network visualization of significant co-occurrences.
For this, we provide the calculation of the co-occurrence significance measures, which we have just introduced, as single function in the file calculateCoocStatistics.R. This function can be imported into the current R-Session with the source command.
# Read in the source code for the co-occurrence calculation
source("calculateCoocStatistics.R")
# Definition of a parameter for the representation of the co-occurrences of a concept
numberOfCoocs <- 15
# Determination of the term of which co-competitors are to be measured.
coocTerm <- "california"We use the imported function calculateCoocStatistics to calculate the co-occurrences for the target term “california”.
coocs <- calculateCoocStatistics(coocTerm, binDTM, measure="LOGLIK")## Loading required package: slam# Display the numberOfCoocs main terms
print(coocs[1:numberOfCoocs])## oregon mexico coast upper mineral texas
## 262.43281 114.40450 64.35717 52.87705 52.84182 51.69327
## acquisition pacific territories york wisconsin arizona
## 43.99069 40.75336 39.47065 36.87249 34.30973 33.98671
## mines iowa miles
## 33.75958 29.69230 29.15576To acquire an extended semantic environment of the target term, ‘secondary co-occurrence’ terms can be computed for each co-occurrence term of the target term. This results in a graph that can be visualized with special layout algorithms (e.g. Force Directed Graph).
Network graphs can be evaluated and visualized in R with the igraph-package. Any graph object can be created from a three-column data-frame. Each row in that data-frame is a triple. Each triple encodes an edge-information of two nodes (source, sink) and an edge-weight value.
For a term co-occurrence network, each triple consists of the target word, a co-occurring word and the significance of their joint occurrence. We denote the values with from, to, sig.
resultGraph <- data.frame(from = character(), to = character(), sig = numeric(0))The process of gathering the network for the target term runs in two steps. First, we obtain all significant co-occurrence terms for the target term. Second, we obtain all co-occurrences of the co-occurrence terms from step one.
Intermediate results for each term are stored as temporary triples named tmpGraph. With the rbind command (“row bind”, used for concatenation of data-frames) all tmpGraph are appended to the complete network object stored in resultGraph.
# The structure of the temporary graph object is equal to that of the resultGraph
tmpGraph <- data.frame(from = character(), to = character(), sig = numeric(0))
# Fill the data.frame to produce the correct number of lines
tmpGraph[1:numberOfCoocs, 3] <- coocs[1:numberOfCoocs]
# Entry of the search word into the first column in all lines
tmpGraph[, 1] <- coocTerm
# Entry of the co-occurrences into the second column of the respective line
tmpGraph[, 2] <- names(coocs)[1:numberOfCoocs]
# Set the significances
tmpGraph[, 3] <- coocs[1:numberOfCoocs]
# Attach the triples to resultGraph
resultGraph <- rbind(resultGraph, tmpGraph)
# Iteration over the most significant numberOfCoocs co-occurrences of the search term
for (i in 1:numberOfCoocs){
# Calling up the co-occurrence calculation for term i from the search words co-occurrences
newCoocTerm <- names(coocs)[i]
coocs2 <- calculateCoocStatistics(newCoocTerm, binDTM, measure="LOGLIK")
#print the co-occurrences
coocs2[1:10]
# Structure of the temporary graph object
tmpGraph <- data.frame(from = character(), to = character(), sig = numeric(0))
tmpGraph[1:numberOfCoocs, 3] <- coocs2[1:numberOfCoocs]
tmpGraph[, 1] <- newCoocTerm
tmpGraph[, 2] <- names(coocs2)[1:numberOfCoocs]
tmpGraph[, 3] <- coocs2[1:numberOfCoocs]
#Append the result to the result graph
resultGraph <- rbind(resultGraph, tmpGraph[2:length(tmpGraph[, 1]), ])
}As a result, resultGraph now contains all numberOfCoocs * numberOfCoocs edges of a term co-occurrence network.
# Sample of some examples from resultGraph
resultGraph[sample(nrow(resultGraph), 6), ]## from to sig
## 1012 arizona pursuit 12.27342
## 93 coast eastern 79.07697
## 138 pacific japan 79.06188
## 1014 iowa kentucky 31.11145
## 613 mines products 35.44232
## 1312 arizona governor 10.21321The package iGraph offers multiple graph visualizations for graph objects. Graph objects can be created from triple lists, such as those we just generated. In the next step we load the package iGraph and create a visualization of all nodes and edges from the object resultGraph.
require(igraph)
# Set the graph and type. In this case, "F" means "Force Directed"
graphNetwork <- graph.data.frame(resultGraph, directed = F)
# Identification of all nodes with less than 2 edges
graphVs <- V(graphNetwork)[degree(graphNetwork) < 2]
# These edges are removed from the graph
graphNetwork <- delete.vertices(graphNetwork, graphVs)
# Assign colors to edges and nodes (searchterm blue, rest orange)
V(graphNetwork)$color <- ifelse(V(graphNetwork)$name == coocTerm, 'cornflowerblue', 'orange')
# Edges with a significance of at least 50% of the maximum sig- nificance in the graph are drawn in orange
halfMaxSig <- max(E(graphNetwork)$sig) * 0.5
E(graphNetwork)$color <- ifelse(E(graphNetwork)$sig > halfMaxSig, "coral", "azure3")
# Disable edges with radius
E(graphNetwork)$curved <- 0
# Size the nodes by their degree of networking
V(graphNetwork)$size <- log(degree(graphNetwork)) * 5
# All nodes must be assigned a standard minimum-size
V(graphNetwork)$size[V(graphNetwork)$size < 5] <- 3
# edge thickness
E(graphNetwork)$width <- 2
# Define the frame and spacing for the plot
par(mai=c(0,0,1,0))
# Finaler Plot
plot(graphNetwork,
layout = layout.fruchterman.reingold, # Force Directed Layout
main = paste(coocTerm, ' Graph'),
vertex.label.family = "sans",
vertex.label.cex = 0.8,
vertex.shape = "circle",
vertex.label.dist = 0.5, # Labels of the nodes moved slightly
vertex.frame.color = 'darkolivegreen',
vertex.label.color = 'black', # Color of node names
vertex.label.font = 2, # Font of node names
vertex.label = V(graphNetwork)$name, # node names
vertex.label.cex = 1 # font size of node names
)
5 Optional exercises
- Create term networks for “civil”, “germany”, “tax”
For visualization, at one point we filter for all nodes with less than 2 edges. By this, the network plot gets less dense, but we loose also a lot of co-occurring terms connected only to one term. Re-draw the network without this filtering.
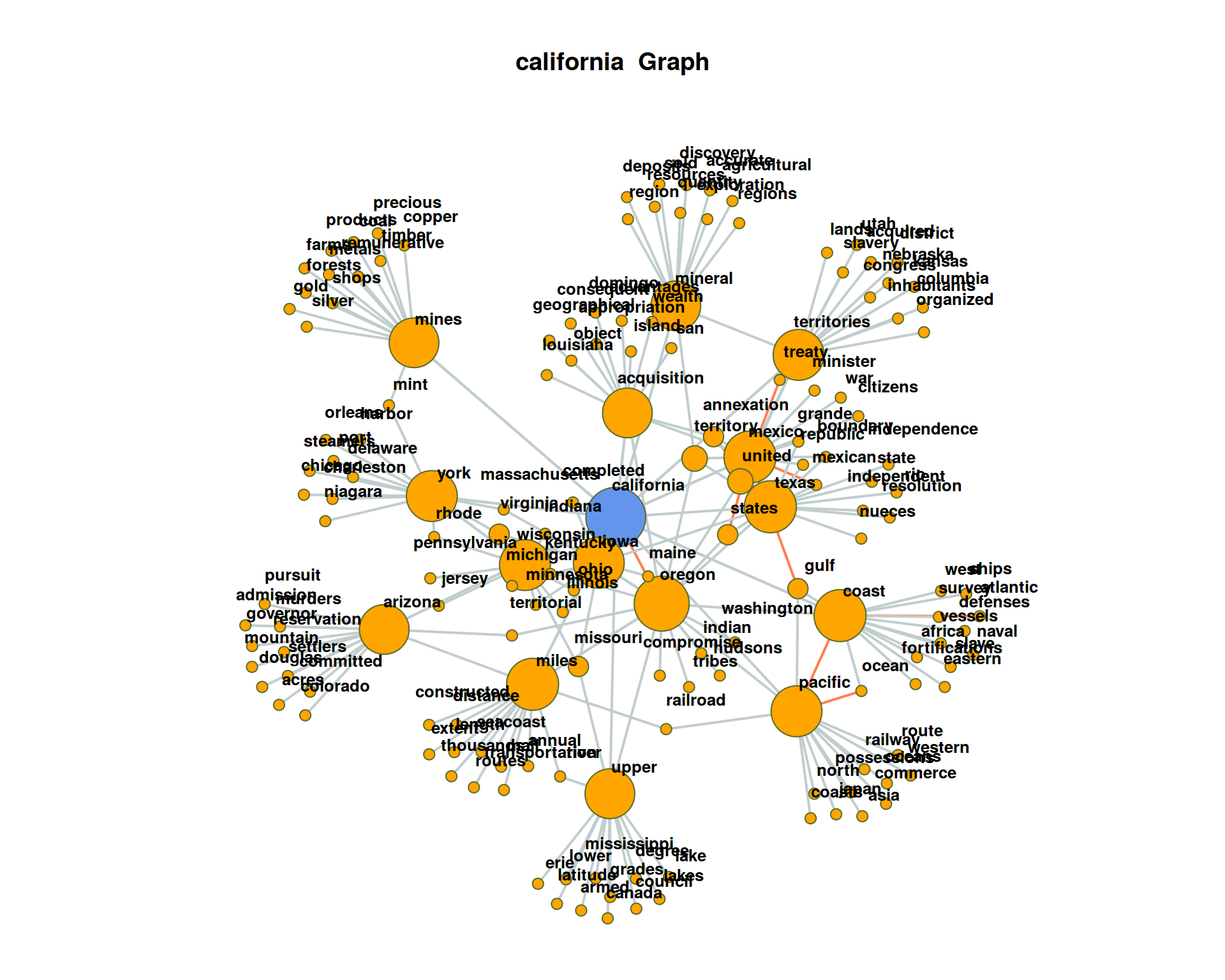 The plot may get very messy. Try lower values for
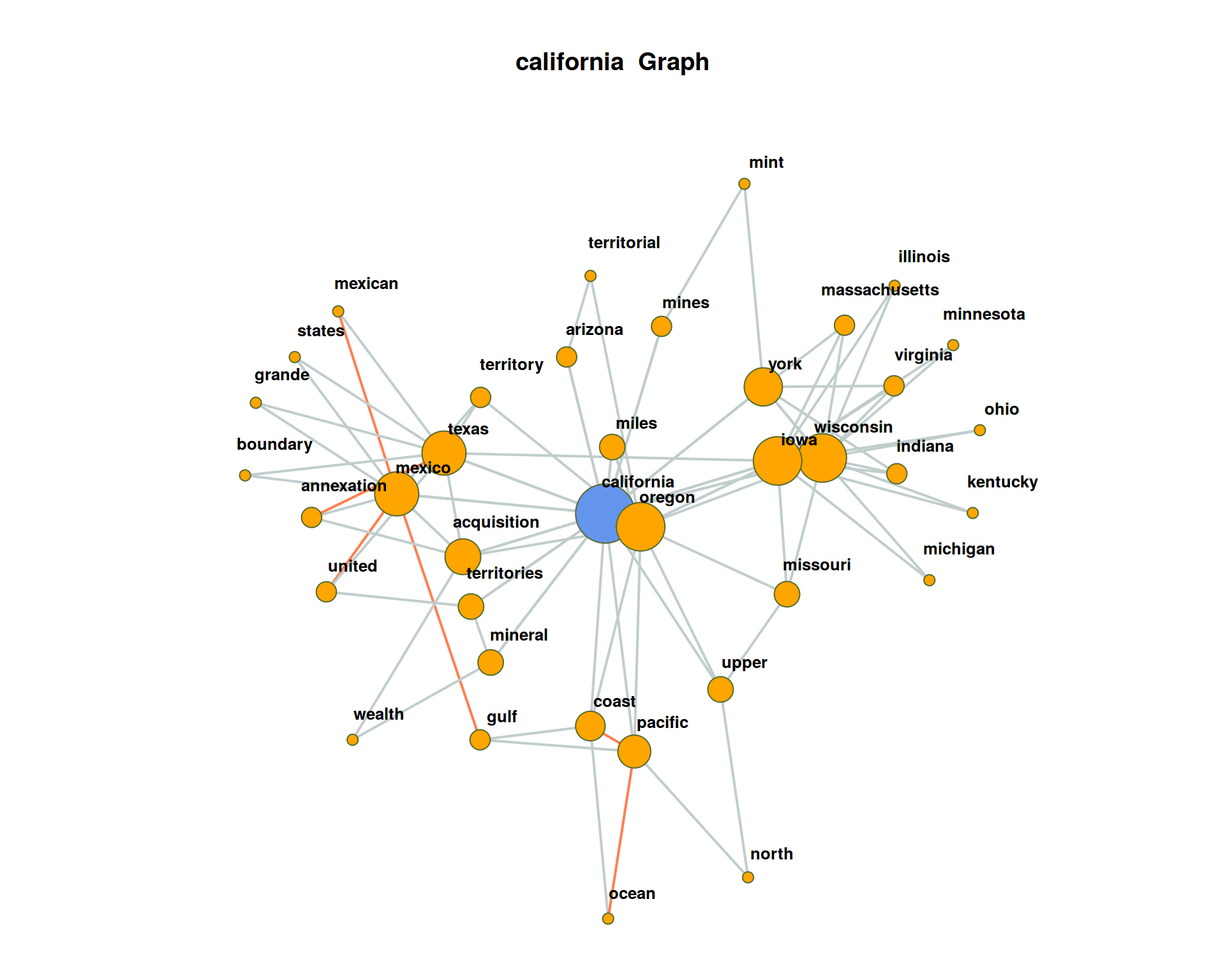
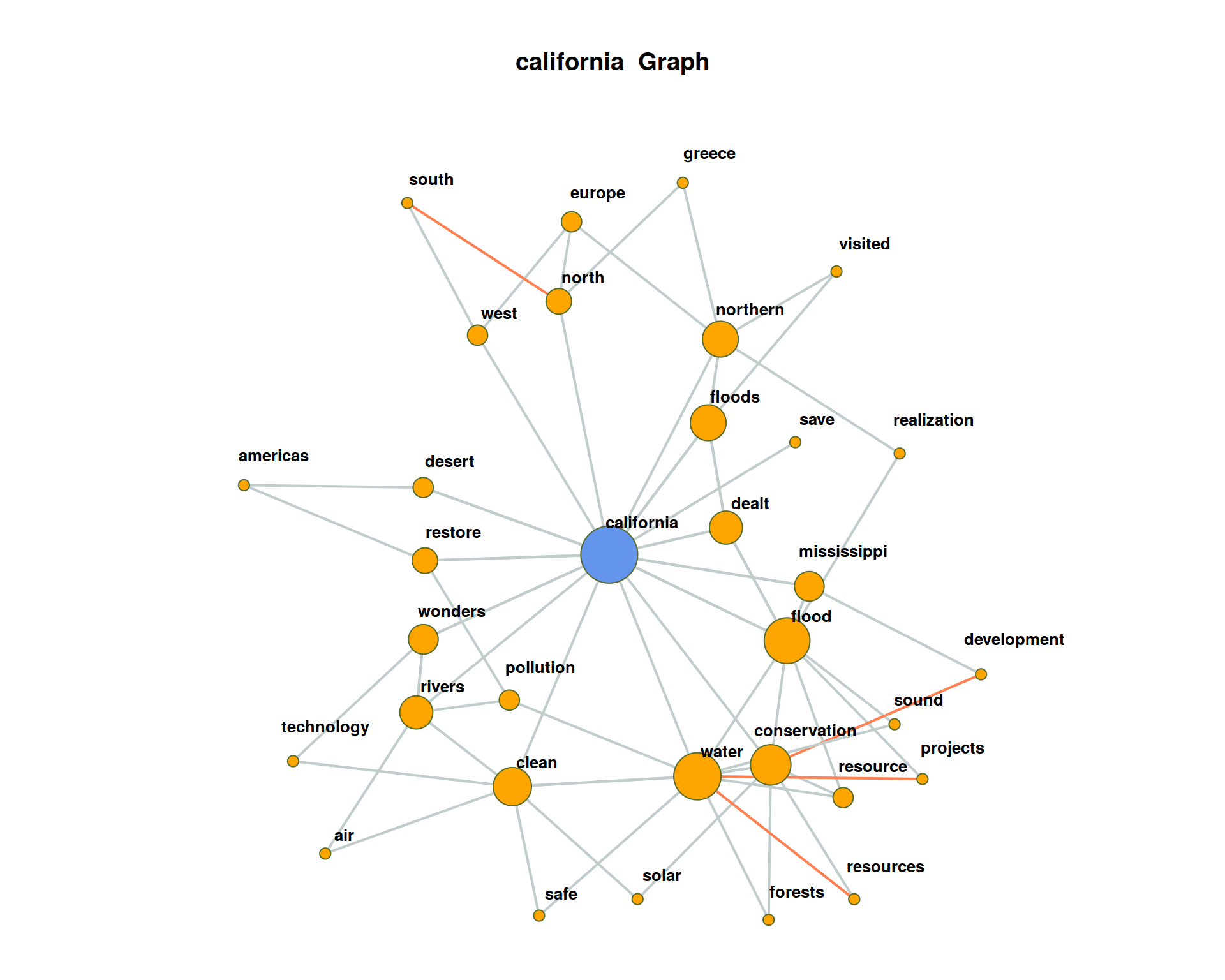
The plot may get very messy. Try lower values for numberOfCoocsto create a less dense network plot.Separate the DTM into two time periods (year < 1950; year > = 1950). Represent the graphs for the term “california” for both time periods. Hint: Define functions for the sub processes of creating a binary DTM from a corpus object (
get_binDTM <- function(mycorpus)) and for visualizing a co-occurrence network (vis_cooc_network <- function(binDTM, coocTerm)).
## ===========================================================================## ===========================================================================
2017, Andreas Niekler and Gregor Wiedemann. GPLv3. tm4ss.github.io